Chapter 21 Causation
Some of these notes are adapted from this tutorial: Mediation and moderation
21.1 Learning goals
- Understanding what controlling for variables means.
- Learning a graphical procedure that helps identify when it’s good vs. bad to control for variables.
- Simulating a mediation analysis.
- Baron and Kenny’s (1986) steps for mediation.
- Testing the significance of a mediation.
- Sobel test.
- Bootstrapping.
- Bayesian approach.
- Limitations of mediation analysis.
- Simulating a moderator effect.
21.3 Load packages and set plotting theme
library("knitr") # for knitting RMarkdown
library("kableExtra") # for making nice tables
library("janitor") # for cleaning column names
library("mediation") # for mediation and moderation analysis
library("multilevel") # Sobel test
library("broom") # tidying up regression results
library("DiagrammeR") # for drawing diagrams
library("DiagrammeRsvg") # for exporting pdfs of graphs
library("rsvg") # for exporting pdfs of graphs
library("tidyverse") # for wrangling, plotting, etc. 21.4 Bayesian networks
21.4.1 Sprinkler example
# cloudy
df.cloudy = tibble(`p(C)` = 0.5)
df.cloudy %>%
kable() %>%
kable_styling(bootstrap_options = "striped",
full_width = F,
font_size = 20)| p(C) |
|---|
| 0.5 |
# sprinkler given cloudy
df.sprinkler_given_cloudy = tibble(C = c("F", "T"),
`p(S)`= c(0.5, 0.1))
df.sprinkler_given_cloudy %>%
kable() %>%
kable_styling(bootstrap_options = "striped",
full_width = F,
font_size = 20)| C | p(S) |
|---|---|
| F | 0.5 |
| T | 0.1 |
# rain given cloudy
df.rain_given_cloudy = tibble(C = c("F", "T"),
`p(R)`= c(0.2, 0.8))
df.rain_given_cloudy %>%
kable() %>%
kable_styling(bootstrap_options = "striped",
full_width = F,
font_size = 20)| C | p(R) |
|---|---|
| F | 0.2 |
| T | 0.8 |
# wet given sprinkler and rain
df.rain_given_sprinkler_and_rain = tibble(
S = rep(c("F", "T"), 2),
R = rep(c("F", "T"), each = 2),
`p(W)`= c(0, 0.9, 0.9, 0.99)
)
df.rain_given_sprinkler_and_rain %>%
kable() %>%
kable_styling(bootstrap_options = "striped",
full_width = F,
font_size = 20)| S | R | p(W) |
|---|---|---|
| F | F | 0.00 |
| T | F | 0.90 |
| F | T | 0.90 |
| T | T | 0.99 |
21.5 Controlling for variables
21.5.1 Illustration of the d-separation algorithm
- Question: Are D and E independent?
21.5.1.1 Full DAG
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
a [label = 'A' pos = '0,0!']
b [label = 'B' pos = '2,0!']
c [label = 'C' pos = '1,-1!']
d [label = 'D' pos = '0,-2!']
e [label = 'E' pos = '2,-2!']
f [label = 'F' pos = '1,-3!']
g [label = 'G' pos = '0,-4!']
# edges between nodes
edge [color = black]
a -> c
b -> c
c -> {d e}
d -> f
f -> g
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/dag.pdf")
# show plot
g21.5.1.2 Draw the ancestral graph
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
a [label = 'A' pos = '0,0!']
b [label = 'B' pos = '2,0!']
c [label = 'C' pos = '1,-1!']
d [label = 'D' pos = '0,-2!']
e [label = 'E' pos = '2,-2!']
# edges between nodes
edge [color = black]
a -> c
b -> c
c -> {d e}
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/ancestral_graph.pdf")
# show plot
g21.5.1.3 “Moralize” the ancestral graph by “marrying” any parents, and disorient by replacing arrows with edges
g = grViz("
graph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
a [label = 'A' pos = '0,0!']
b [label = 'B' pos = '2,0!']
c [label = 'C' pos = '1,-1!']
d [label = 'D' pos = '0,-2!']
e [label = 'E' pos = '2,-2!']
# edges between nodes
edge [color = black]
a -- c
b -- c
c -- {d e}
edge [color = black]
a -- b
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/moralize_and_disorient.pdf")
# show plot
g- For the case in which we check whether D and E are independent conditioned on C
g = grViz("
graph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
a [label = 'A' pos = '0,0!']
b [label = 'B' pos = '2,0!']
d [label = 'D' pos = '0,-2!']
e [label = 'E' pos = '2,-2!']
# edges between nodes
edge [color = black]
edge [color = black]
a -- b
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
## export as pdf
#g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/moralize_and_disorient2.pdf")
# show plot
g21.5.2 Good controls
21.5.2.1 Common cause (with direct link between X and Y)
21.5.2.1.1 DAG
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '2,0!']
z [label = 'Z' pos = '1,1!', fontcolor = 'red']
# edges between nodes
edge [color = black]
x -> y
z -> {x y}
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/common_cause1.pdf")
# show plot
g21.5.2.1.2 Regression
set.seed(1)
n = 1000
b_zx = 2
b_xy = 2
b_zy = 2
sd = 1
df = tibble(z = rnorm(n = n, sd = sd),
x = b_zx * z + rnorm(n = n, sd = sd),
y = b_zy * z + b_xy * x + rnorm(n = n, sd = sd))
# without control
lm(formula = y ~ x,
data = df) %>%
summary()
Call:
lm(formula = y ~ x, data = df)
Residuals:
Min 1Q Median 3Q Max
-4.6011 -0.9270 -0.0506 0.9711 4.0454
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.02449 0.04389 0.558 0.577
x 2.82092 0.01890 149.225 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.388 on 998 degrees of freedom
Multiple R-squared: 0.9571, Adjusted R-squared: 0.9571
F-statistic: 2.227e+04 on 1 and 998 DF, p-value: < 2.2e-16
Call:
lm(formula = y ~ x + z, data = df)
Residuals:
Min 1Q Median 3Q Max
-3.6151 -0.6564 -0.0223 0.6815 2.8132
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.01624 0.03260 0.498 0.618
x 2.02202 0.03135 64.489 <2e-16 ***
z 2.00501 0.07036 28.497 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.031 on 997 degrees of freedom
Multiple R-squared: 0.9764, Adjusted R-squared: 0.9763
F-statistic: 2.059e+04 on 2 and 997 DF, p-value: < 2.2e-1621.5.2.1.3 Moralize and disorient the ancestral graph
g = grViz("
graph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '2,0!']
z [label = 'Z' pos = '1,1!', fontcolor = 'red']
# edges between nodes
edge [color = black]
x -- y
z -- {x y}
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/common_cause1_undirected.pdf")
# # rsvg_pdf("figures/common_cause1_undirected2.pdf")
# show plot
g21.5.2.2 Common effect (with direct link between X and Y)#### Common cause (without direct link between X and Y)
21.5.2.2.1 DAG
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '2,0!']
z [label = 'Z' pos = '1,1!', fontcolor = 'red']
# edges between nodes
edge [color = black]
z -> {x y}
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/common_cause2.pdf")
# show plot
g21.5.2.2.2 Regression
set.seed(1)
n = 1000
b_zx = 2
b_zy = 2
sd = 1
df = tibble(z = rnorm(n = n, sd = sd),
x = b_zx * z + rnorm(n = n, sd = sd),
y = b_zy * z + rnorm(n = n, sd = sd))
# without control
lm(formula = y ~ x,
data = df) %>%
summary()
Call:
lm(formula = y ~ x, data = df)
Residuals:
Min 1Q Median 3Q Max
-4.6011 -0.9270 -0.0506 0.9711 4.0454
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.02449 0.04389 0.558 0.577
x 0.82092 0.01890 43.426 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.388 on 998 degrees of freedom
Multiple R-squared: 0.6539, Adjusted R-squared: 0.6536
F-statistic: 1886 on 1 and 998 DF, p-value: < 2.2e-16
Call:
lm(formula = y ~ x + z, data = df)
Residuals:
Min 1Q Median 3Q Max
-3.6151 -0.6564 -0.0223 0.6815 2.8132
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.01624 0.03260 0.498 0.618
x 0.02202 0.03135 0.702 0.483
z 2.00501 0.07036 28.497 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.031 on 997 degrees of freedom
Multiple R-squared: 0.8093, Adjusted R-squared: 0.8089
F-statistic: 2115 on 2 and 997 DF, p-value: < 2.2e-1621.5.3 Bad controls
21.5.3.1 Common effect
21.5.3.1.1 DAG
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '2,0!']
z [label = 'Z' pos = '1,-1!', fontcolor = 'red']
# edges between nodes
edge [color = black]
x -> z
y -> z
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/common_effect.pdf")
# show plot
g21.5.3.1.2 Regression
set.seed(1)
n = 1000
b_xz = 2
b_yz = 2
sd = 1
df = tibble(x = rnorm(n = n, sd = sd),
y = rnorm(n = n, sd = sd),
z = x * b_xz + y * b_yz + rnorm(n = n, sd = sd))
# without control
lm(formula = y ~ x,
data = df) %>%
summary()
Call:
lm(formula = y ~ x, data = df)
Residuals:
Min 1Q Median 3Q Max
-3.2484 -0.6720 -0.0138 0.7554 3.6443
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.016187 0.032905 -0.492 0.623
x 0.006433 0.031809 0.202 0.840
Residual standard error: 1.04 on 998 degrees of freedom
Multiple R-squared: 4.098e-05, Adjusted R-squared: -0.000961
F-statistic: 0.0409 on 1 and 998 DF, p-value: 0.8398
Call:
lm(formula = y ~ x + z, data = df)
Residuals:
Min 1Q Median 3Q Max
-1.35547 -0.30016 0.00298 0.31119 1.73408
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.009608 0.014477 -0.664 0.507
x -0.816164 0.018936 -43.102 <2e-16 ***
z 0.398921 0.006186 64.489 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4578 on 997 degrees of freedom
Multiple R-squared: 0.8066, Adjusted R-squared: 0.8062
F-statistic: 2079 on 2 and 997 DF, p-value: < 2.2e-1621.5.3.1.3 Moralize and disorient the ancestral graph
g = grViz("
graph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '2,0!']
z [label = 'Z' pos = '1,-1!', fontcolor = 'red']
# edges between nodes
edge [color = black]
x -- y
x -- z
y -- z
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/common_effect_undirected1.pdf")
# rsvg_pdf("figures/common_effect_undirected2.pdf")
# show plot
g21.5.3.2 Causal chain 1
21.5.3.2.1 DAG
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '2,0!']
z [label = 'Z' pos = '1, 0!', fontcolor = 'red']
# edges between nodes
edge [color = black]
x -> z
z -> y
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# # export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/causal_chain.pdf")
# show plot
g21.5.3.2.2 Regression
set.seed(1)
n = 20
b_xz = 2
b_zy = 2
sd = 1
df = tibble(x = rnorm(n = n, sd = sd),
z = x * b_xz + rnorm(n = n, sd = sd),
y = z * b_zy + rnorm(n = n, sd = sd))
# without control
lm(formula = y ~ x,
data = df) %>%
summary()
Call:
lm(formula = y ~ x, data = df)
Residuals:
Min 1Q Median 3Q Max
-3.336 -1.208 0.209 1.220 3.189
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.1547 0.3982 0.388 0.702
x 3.8488 0.4374 8.799 6.15e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.741 on 18 degrees of freedom
Multiple R-squared: 0.8114, Adjusted R-squared: 0.8009
F-statistic: 77.43 on 1 and 18 DF, p-value: 6.154e-08
Call:
lm(formula = y ~ x + z, data = df)
Residuals:
Min 1Q Median 3Q Max
-1.35959 -0.56643 -0.06193 0.48088 1.80592
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.09559 0.18177 0.526 0.606
x 0.64724 0.43278 1.496 0.153
z 1.78614 0.21425 8.337 2.07e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.7943 on 17 degrees of freedom
Multiple R-squared: 0.9629, Adjusted R-squared: 0.9586
F-statistic: 220.8 on 2 and 17 DF, p-value: 6.868e-1321.5.3.3 Causal chain 2
21.5.3.3.1 DAG
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '1,0!']
z [label = 'Z' pos = '2, 0!', fontcolor = 'red']
# edges between nodes
edge [color = black]
x -> y
y -> z
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# # export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/causal_chain2.pdf")
# show plot
g21.5.3.3.2 Regression
set.seed(1)
n = 20
b_xy = 2
b_yz = 2
sd = 1
df = tibble(x = rnorm(n = n, sd = sd),
y = x * b_xy + rnorm(n = n, sd = sd),
z = y * b_yz + rnorm(n = n, sd = sd),)
# without control
lm(formula = y ~ x,
data = df) %>%
summary()
Call:
lm(formula = y ~ x, data = df)
Residuals:
Min 1Q Median 3Q Max
-1.69133 -0.43739 -0.07132 0.68033 1.63937
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.03307 0.19981 0.166 0.87
x 1.79245 0.21951 8.166 1.83e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.8738 on 18 degrees of freedom
Multiple R-squared: 0.7874, Adjusted R-squared: 0.7756
F-statistic: 66.68 on 1 and 18 DF, p-value: 1.827e-07
Call:
lm(formula = y ~ x + z, data = df)
Residuals:
Min 1Q Median 3Q Max
-0.9023 -0.2316 0.1173 0.2396 0.6319
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.03650 0.09153 -0.399 0.695
x 0.06113 0.23056 0.265 0.794
z 0.44983 0.05396 8.337 2.07e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.3986 on 17 degrees of freedom
Multiple R-squared: 0.9582, Adjusted R-squared: 0.9533
F-statistic: 195 on 2 and 17 DF, p-value: 1.896e-1221.5.3.4 Bias amplification
21.5.3.4.1 DAG
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
y [label = 'Y' pos = '2,0!']
z [label = 'Z' pos = '-1, 1!', fontcolor = 'red']
u [label = 'U' pos = '1, 1!', fillcolor = 'white']
# edges between nodes
edge [color = black]
x -> y
z -> x
u -> {x y}
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
# # export as pdf
# g %>%
# export_svg %>%
# charToRaw %>%
# rsvg_pdf("figures/bias_amplification.pdf")
# show plot
g21.5.3.5 Regression
set.seed(1)
n = 20
b_xy = 2
b_ux = 2
b_uy = 2
b_zx = 2
sd = 1
df = tibble(u = rnorm(n = n, sd = sd),
z = rnorm(n = n, sd = sd),
x = u * b_ux + z * b_zx + rnorm(n = n, sd = sd),
y = u * b_uy + x * b_xy + rnorm(n = n, sd = sd))
# without control
lm(formula = y ~ x,
data = df) %>%
summary()
Call:
lm(formula = y ~ x, data = df)
Residuals:
Min 1Q Median 3Q Max
-2.2838 -0.8662 -0.2281 0.7201 3.1619
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.1903 0.3282 0.58 0.569
x 2.5771 0.1375 18.74 2.96e-13 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.434 on 18 degrees of freedom
Multiple R-squared: 0.9512, Adjusted R-squared: 0.9485
F-statistic: 351.1 on 1 and 18 DF, p-value: 2.961e-13
Call:
lm(formula = y ~ x + z, data = df)
Residuals:
Min 1Q Median 3Q Max
-1.9114 -0.4876 -0.1044 0.6333 1.8935
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.07431 0.24564 0.303 0.76594
x 2.78984 0.11553 24.147 1.35e-14 ***
z -1.25270 0.31719 -3.949 0.00103 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.066 on 17 degrees of freedom
Multiple R-squared: 0.9746, Adjusted R-squared: 0.9716
F-statistic: 325.7 on 2 and 17 DF, p-value: 2.793e-1421.6 Mediation
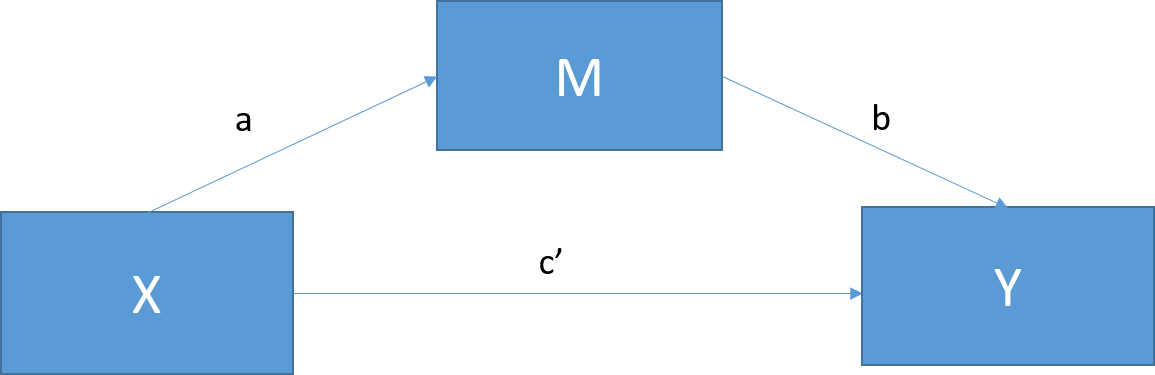
Figure 21.1: Basic mediation model. c = the total effect of X on Y; c = c’ + ab; c’ = the direct effect of X on Y after controlling for M; c’ = c - ab; ab = indirect effect of X on Y.
Mediation tests whether the effects of X (the independent variable) on Y (the dependent variable) operate through a third variable, M (the mediator). In this way, mediators explain the causal relationship between two variables or “how” the relationship works, making it a very popular method in psychological research.
Figure 21.1 shows the standard mediation model. Perfect mediation occurs when the effect of X on Y decreases to 0 with M in the model. Partial mediation occurs when the effect of X on Y decreases by a nontrivial amount (the actual amount is up for debate) with M in the model.
Important: Both mediation and moderation assume that the DV did not CAUSE the mediator/moderator.
21.6.2 Method 1: Baron & Kenny’s (1986) indirect effect method
The Baron and Kenny (1986) method is among the original methods for testing for mediation but tends to have low statistical power.
The three steps:
Estimate the relationship between \(X\) and \(Y\) (hours since dawn on degree of wakefulness). Path “c” must be significantly different from 0; must have a total effect between the IV & DV.
Estimate the relationship between \(X\) and \(M\) (hours since dawn on coffee consumption). Path “a” must be significantly different from 0; IV and mediator must be related.
Estimate the relationship between \(M\) and \(Y\) controlling for \(X\) (coffee consumption on wakefulness, controlling for hours since dawn). Path “b” must be significantly different from 0; mediator and DV must be related. The effect of \(X\) on \(Y\) decreases with the inclusion of \(M\) in the model.
21.6.2.1 Total effect
Total effect of X on Y (not controlling for M).
# fit the model
fit.y_x = lm(formula = y ~ 1 + x,
data = df.mediation)
# summarize the results
fit.y_x %>%
summary()
Call:
lm(formula = y ~ 1 + x, data = df.mediation)
Residuals:
Min 1Q Median 3Q Max
-10.917 -3.738 -0.259 2.910 12.540
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 8.78300 6.16002 1.426 0.1571
x 0.16899 0.08116 2.082 0.0399 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.16 on 98 degrees of freedom
Multiple R-squared: 0.04237, Adjusted R-squared: 0.0326
F-statistic: 4.336 on 1 and 98 DF, p-value: 0.0399321.6.2.2 Path a
Call:
lm(formula = m ~ 1 + x, data = df.mediation)
Residuals:
Min 1Q Median 3Q Max
-9.5367 -3.4175 -0.4375 2.9032 16.4520
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.29696 5.79432 0.396 0.693
x 0.66252 0.07634 8.678 8.87e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 4.854 on 98 degrees of freedom
Multiple R-squared: 0.4346, Adjusted R-squared: 0.4288
F-statistic: 75.31 on 1 and 98 DF, p-value: 8.872e-1421.6.2.3 Path b and c’
Effect of M on Y controlling for X.
Call:
lm(formula = y ~ 1 + m + x, data = df.mediation)
Residuals:
Min 1Q Median 3Q Max
-9.3651 -3.3037 -0.6222 3.1068 10.3991
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.80952 5.68297 1.374 0.173
m 0.42381 0.09899 4.281 4.37e-05 ***
x -0.11179 0.09949 -1.124 0.264
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 4.756 on 97 degrees of freedom
Multiple R-squared: 0.1946, Adjusted R-squared: 0.1779
F-statistic: 11.72 on 2 and 97 DF, p-value: 2.771e-0521.6.2.4 Interpretation
fit.y_x %>%
tidy() %>%
mutate(path = "c") %>%
bind_rows(fit.m_x %>%
tidy() %>%
mutate(path = "a"),
fit.y_mx %>%
tidy() %>%
mutate(path = c("(Intercept)", "b", "c'"))) %>%
filter(term != "(Intercept)") %>%
mutate(significance = p.value < .05,
dv = ifelse(path %in% c("c'", "b"), "y", "m")) %>%
select(path, iv = term, dv, estimate, p.value, significance)# A tibble: 4 × 6
path iv dv estimate p.value significance
<chr> <chr> <chr> <dbl> <dbl> <lgl>
1 c x m 0.169 3.99e- 2 TRUE
2 a x m 0.663 8.87e-14 TRUE
3 b m y 0.424 4.37e- 5 TRUE
4 c' x y -0.112 2.64e- 1 FALSE Here we find that our total effect model shows a significant positive relationship between hours since dawn (X) and wakefulness (Y). Our Path A model shows that hours since down (X) is also positively related to coffee consumption (M). Our Path B model then shows that coffee consumption (M) positively predicts wakefulness (Y) when controlling for hours since dawn (X).
Since the relationship between hours since dawn and wakefulness is no longer significant when controlling for coffee consumption, this suggests that coffee consumption does in fact mediate this relationship. However, this method alone does not allow for a formal test of the indirect effect so we don’t know if the change in this relationship is truly meaningful.
21.6.3 Method 2: Sobel Test
The Sobel Test tests whether the indirect effect from X via M to Y is significant.
# run the sobel test
fit.sobel = sobel(pred = df.mediation$x,
med = df.mediation$m,
out = df.mediation$y)
# calculate the p-value
(1 - pnorm(fit.sobel$z.value))*2[1] 0.0001233403The relationship between “hours since dawn” and “wakefulness” is significantly mediated by “coffee consumption”.
The Sobel Test is largely considered an outdated method since it assumes that the indirect effect (ab) is normally distributed and tends to only have adequate power with large sample sizes. Thus, again, it is highly recommended to use the mediation bootstrapping method instead.
21.6.4 Method 3: Bootstrapping
The “mediation” packages uses the more recent bootstrapping method of Preacher and Hayes (2004) to address the power limitations of the Sobel Test.
This method does not require that the data are normally distributed, and is particularly suitable for small sample sizes.
# bootstrapped mediation
fit.mediation = mediate(model.m = fit.m_x,
model.y = fit.y_mx,
treat = "x",
mediator = "m",
boot = T)Running nonparametric bootstrap
Causal Mediation Analysis
Nonparametric Bootstrap Confidence Intervals with the Percentile Method
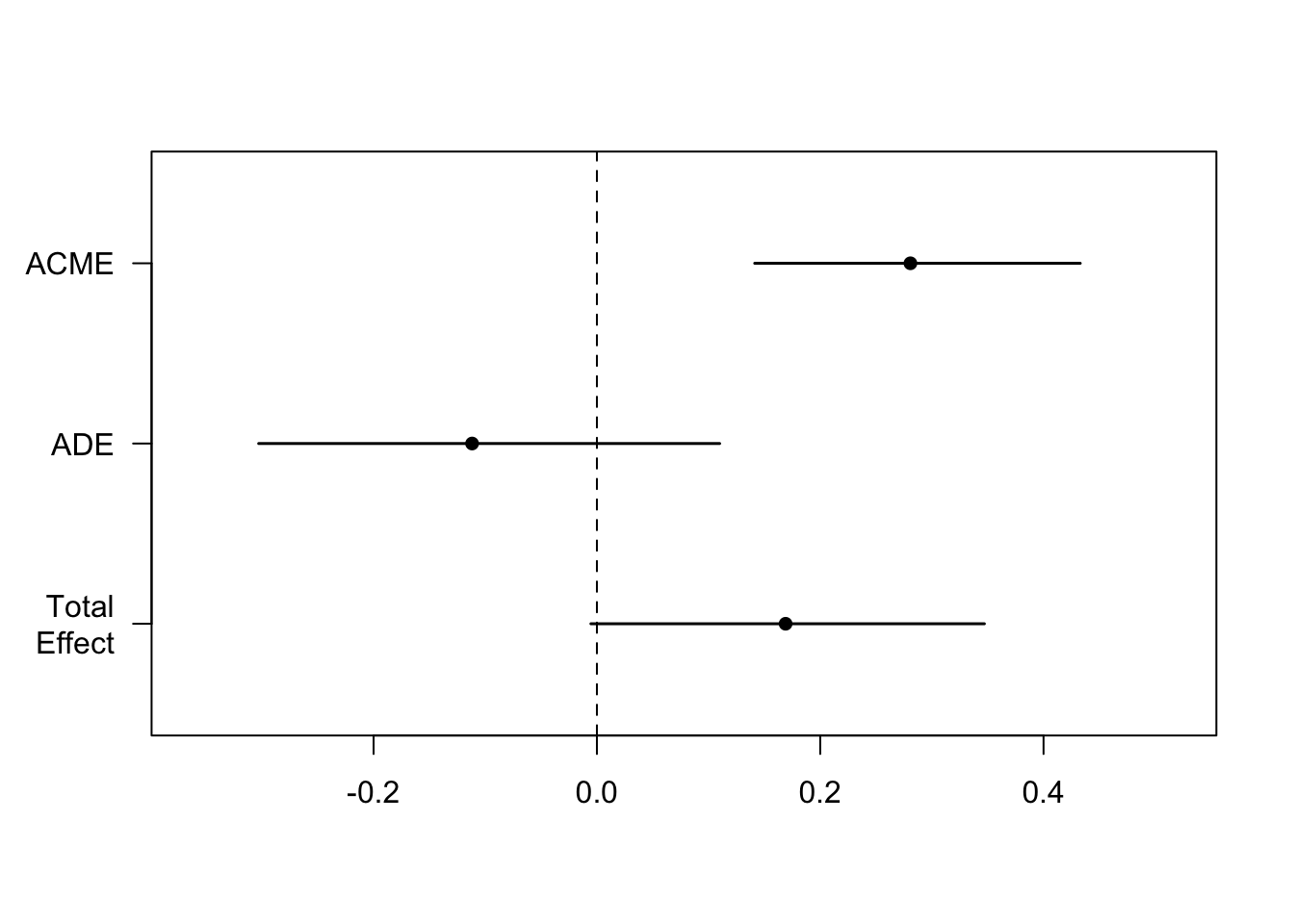
Estimate 95% CI Lower 95% CI Upper p-value
ACME 0.28078 0.14133 0.43 <2e-16 ***
ADE -0.11179 -0.30293 0.11 0.274
Total Effect 0.16899 -0.00539 0.35 0.056 .
Prop. Mediated 1.66151 -1.91644 10.16 0.056 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 100
Simulations: 1000 - ACME = Average causal mediation effect
- ADE = Average direct effect
- Total effect = ACME + ADE
Plot the results:
21.6.4.1 Interpretation
The mediate() function gives us our Average Causal Mediation Effects (ACME), our Average Direct Effects (ADE), our combined indirect and direct effects (Total Effect), and the ratio of these estimates (Prop. Mediated). The ACME here is the indirect effect of M (total effect - direct effect) and thus this value tells us if our mediation effect is significant.
21.6.5 Be careful about mediation!
Different causal structures can lead to the same “mediation” effects. So it’s difficult to tell from a pattern of regression results what the relationships between variables is.
Here are three different causal structures that lead to similar effects
set.seed(1)
n = 100 # number of observations
# causal chain
df.causal_chain = tibble(x = rnorm(n, 0, 1),
z = 2 * x + rnorm(n, 0, 1),
y = 2 * z + rnorm(n, 0, 1))
# visualize the graph
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
z [label = 'Z' pos = '1,1!']
y [label = 'Y' pos = '2,0!']
# edges between nodes
edge [color = black]
x -> z
z -> y
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
g# fit models
fit.yx = lm(formula = y ~ 1 + x,
data = df.causal_chain)
fit.zx = lm(formula = z ~ 1 + x,
data = df.causal_chain)
fit.yxz = lm(formula = y ~ 1 + x + z,
data = df.causal_chain)
# print models
summary(fit.yx)
Call:
lm(formula = y ~ 1 + x, data = df.causal_chain)
Residuals:
Min 1Q Median 3Q Max
-4.0180 -1.2143 -0.4065 1.1288 6.0959
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.04802 0.21582 -0.222 0.824
x 4.01905 0.23972 16.766 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.142 on 98 degrees of freedom
Multiple R-squared: 0.7415, Adjusted R-squared: 0.7389
F-statistic: 281.1 on 1 and 98 DF, p-value: < 2.2e-16
Call:
lm(formula = z ~ 1 + x, data = df.causal_chain)
Residuals:
Min 1Q Median 3Q Max
-1.8768 -0.6138 -0.1395 0.5394 2.3462
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.03769 0.09699 -0.389 0.698
x 1.99894 0.10773 18.556 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.9628 on 98 degrees of freedom
Multiple R-squared: 0.7784, Adjusted R-squared: 0.7762
F-statistic: 344.3 on 1 and 98 DF, p-value: < 2.2e-16
Call:
lm(formula = y ~ 1 + x + z, data = df.causal_chain)
Residuals:
Min 1Q Median 3Q Max
-2.94359 -0.43645 0.00202 0.63692 2.63941
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.02535 0.10519 0.241 0.810
x 0.12804 0.24804 0.516 0.607
z 1.94653 0.10948 17.780 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.043 on 97 degrees of freedom
Multiple R-squared: 0.9393, Adjusted R-squared: 0.9381
F-statistic: 750.6 on 2 and 97 DF, p-value: < 2.2e-16# mediation analysis
fit.m = mediate(model.m = fit.zx,
model.y = fit.yxz,
treat = "x",
mediator = "z",
boot = T)Running nonparametric bootstrap
Causal Mediation Analysis
Nonparametric Bootstrap Confidence Intervals with the Percentile Method
Estimate 95% CI Lower 95% CI Upper p-value
ACME 3.891 3.390 4.51 <2e-16 ***
ADE 0.128 -0.395 0.62 0.64
Total Effect 4.019 3.585 4.44 <2e-16 ***
Prop. Mediated 0.968 0.849 1.11 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 100
Simulations: 1000 set.seed(1)
n = 100 # number of observations
# common cause
df.common_cause = tibble(z = rnorm(n, 0, 1),
x = 2 * z + rnorm(n, 0, 1),
y = 2 * z + rnorm(n, 0, 1))
# visualize the graph
g = grViz("
digraph neato {
graph[layout = neato]
# general settings for all nodes
node [
shape = circle,
style = filled,
color = black,
label = ''
fontname = 'Helvetica',
fontsize = 16,
fillcolor = lightblue
]
# labels for each node
x [label = 'X' pos = '0,0!']
z [label = 'Z' pos = '1,1!']
y [label = 'Y' pos = '2,0!']
# edges between nodes
edge [color = black]
z -> x
z -> y
# direction in which arrows are drawn (from left to right)
rankdir = LR
}
")
g# fit models
fit.yx = lm(formula = y ~ 1 + x,
data = df.common_cause)
fit.zx = lm(formula = z ~ 1 + x,
data = df.common_cause)
fit.yxz = lm(formula = y ~ 1 + x + z,
data = df.common_cause)
# print models
summary(fit.yx)
Call:
lm(formula = y ~ 1 + x, data = df.common_cause)
Residuals:
Min 1Q Median 3Q Max
-3.4514 -0.9780 -0.1073 1.0144 3.3204
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.10793 0.13821 0.781 0.437
x 0.77525 0.06799 11.402 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.377 on 98 degrees of freedom
Multiple R-squared: 0.5702, Adjusted R-squared: 0.5658
F-statistic: 130 on 1 and 98 DF, p-value: < 2.2e-16
Call:
lm(formula = z ~ 1 + x, data = df.common_cause)
Residuals:
Min 1Q Median 3Q Max
-0.90848 -0.28101 0.06274 0.24570 0.85736
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.03880 0.04266 0.91 0.365
x 0.38942 0.02099 18.56 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4249 on 98 degrees of freedom
Multiple R-squared: 0.7784, Adjusted R-squared: 0.7762
F-statistic: 344.3 on 1 and 98 DF, p-value: < 2.2e-16
Call:
lm(formula = y ~ 1 + x + z, data = df.common_cause)
Residuals:
Min 1Q Median 3Q Max
-2.94359 -0.43645 0.00202 0.63692 2.63941
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.02535 0.10519 0.241 0.810
x -0.05347 0.10948 -0.488 0.626
z 2.12804 0.24804 8.580 1.55e-13 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.043 on 97 degrees of freedom
Multiple R-squared: 0.7556, Adjusted R-squared: 0.7506
F-statistic: 150 on 2 and 97 DF, p-value: < 2.2e-16# mediation analysis
fit.m = mediate(model.m = fit.zx,
model.y = fit.yxz,
treat = "x",
mediator = "z",
boot = T)Running nonparametric bootstrap
Causal Mediation Analysis
Nonparametric Bootstrap Confidence Intervals with the Percentile Method
Estimate 95% CI Lower 95% CI Upper p-value
ACME 0.8287 0.6065 1.04 <2e-16 ***
ADE -0.0535 -0.2675 0.16 0.56
Total Effect 0.7752 0.6353 0.90 <2e-16 ***
Prop. Mediated 1.0690 0.8134 1.37 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Sample Size Used: 100
Simulations: 1000 Both models here lead to the same pattern of “mediation”. However, only the first one is a chain: X–>Z–>Y. The second one is a common cause X<–Z–>Y (where X does not cause Y).
21.7 Moderation
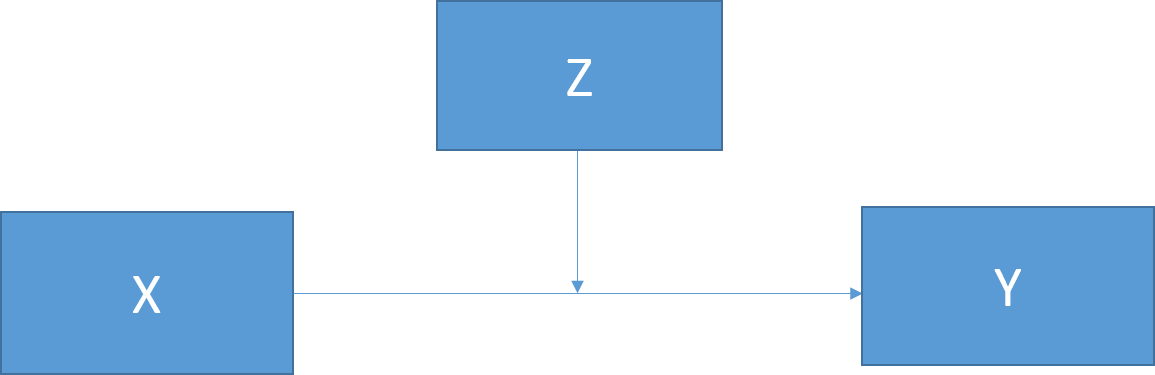
Figure 21.2: Basic moderation model.
Moderation can be tested by looking for significant interactions between the moderating variable (Z) and the IV (X). Notably, it is important to mean center both your moderator and your IV to reduce multicolinearity and make interpretation easier.
21.7.1 Generate data
# make example reproducible
set.seed(123)
# number of participants
n = 100
df.moderation = tibble(x = abs(rnorm(n = n,
mean = 6,
sd = 4)), # hours of sleep
x1 = abs(rnorm(n = n,
mean = 60,
sd = 30)), # adding some systematic variance to our DV
z = rnorm(n = n,
mean = 30,
sd = 8), # ounces of coffee consumed
y = abs((-0.8 * x) * (0.2 * z) - 0.5 * x - 0.4 * x1 + 10 +
rnorm(n, 0, 3))) # attention Paid21.7.2 Moderation analysis
# scale the predictors
df.moderation = df.moderation %>%
mutate(across(.cols = c(x, z),
.fns = ~ scale(.)[,]))
# run regression model with interaction
fit.moderation = lm(formula = y ~ 1 + x * z,
data = df.moderation)
# summarize result
fit.moderation %>%
summary()
Call:
lm(formula = y ~ 1 + x * z, data = df.moderation)
Residuals:
Min 1Q Median 3Q Max
-21.466 -8.972 -0.233 6.180 38.051
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 48.544 1.173 41.390 < 2e-16 ***
x 17.863 1.196 14.936 < 2e-16 ***
z 8.393 1.181 7.108 2.08e-10 ***
x:z 6.094 1.077 5.656 1.59e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 11.65 on 96 degrees of freedom
Multiple R-squared: 0.7661, Adjusted R-squared: 0.7587
F-statistic: 104.8 on 3 and 96 DF, p-value: < 2.2e-1621.7.2.1 Visualize result
# generate data grid with three levels of the moderator
df.newdata = df.moderation %>%
expand(x = c(min(x),
max(x)),
z = c(mean(z) - sd(z),
mean(z),
mean(z) + sd(z))) %>%
mutate(moderator = rep(c("low", "average", "high"), nrow(.)/3))
# predictions for the three levels of the moderator
df.prediction = fit.moderation %>%
augment(newdata = df.newdata) %>%
mutate(moderator = factor(moderator, levels = c("high", "average", "low")))
# visualize the result
df.moderation %>%
ggplot(aes(x = x,
y = y)) +
geom_point() +
geom_line(data = df.prediction,
mapping = aes(y = .fitted,
group = moderator,
color = moderator),
size = 1) +
labs(x = "hours of sleep (z-scored)",
y = "attention paid",
color = "coffee consumed") +
scale_color_brewer(palette = "Set1")Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.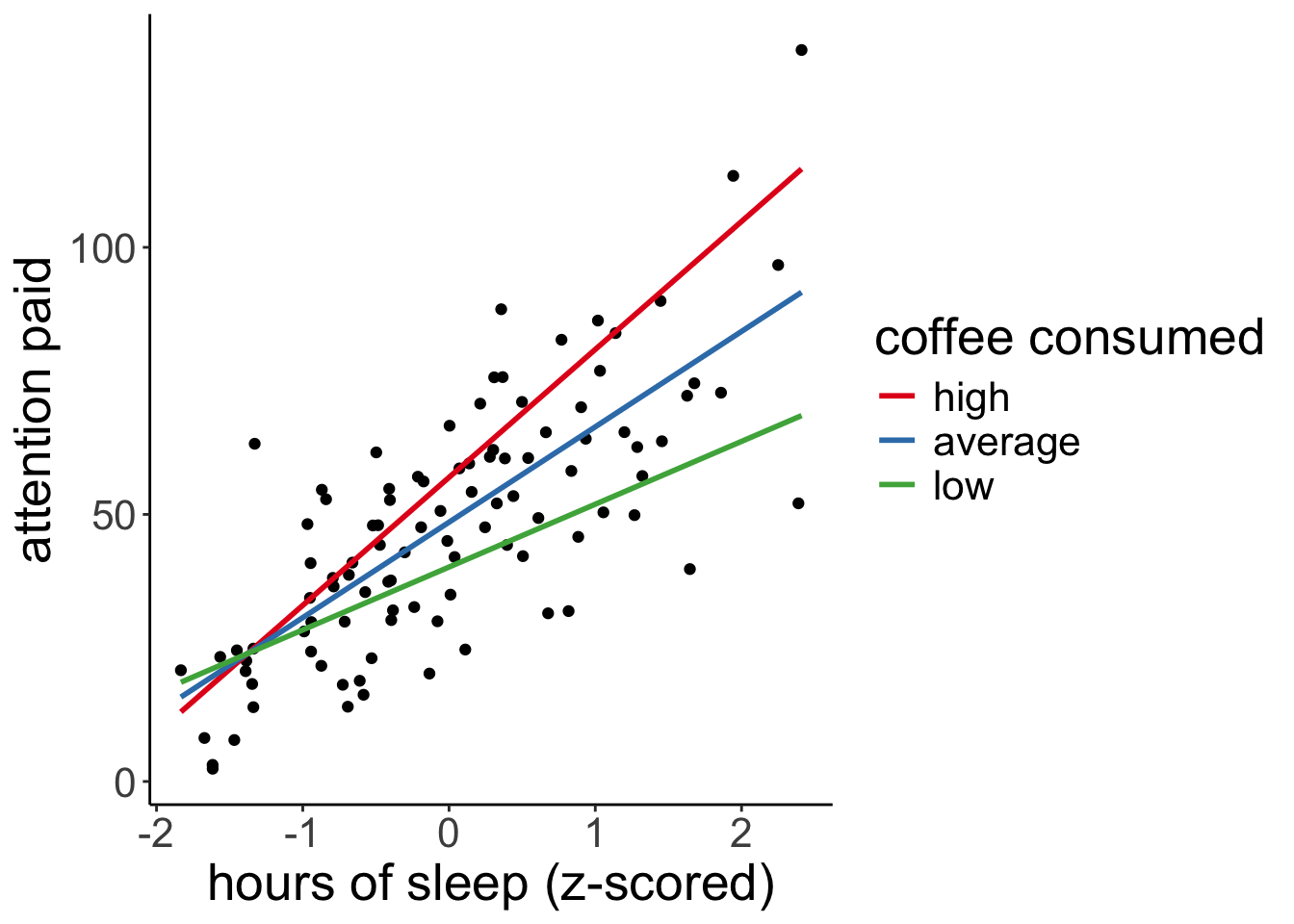
df.prediction %>%
head(9) %>%
kable(digits = 2) %>%
kable_styling(bootstrap_options = "striped",
full_width = F)| x | z | moderator | .fitted |
|---|---|---|---|
| -1.83 | -1 | low | 18.58 |
| -1.83 | 0 | average | 15.80 |
| -1.83 | 1 | high | 13.02 |
| 2.41 | -1 | low | 68.52 |
| 2.41 | 0 | average | 91.60 |
| 2.41 | 1 | high | 114.68 |
21.9 Session info
Information about this R session including which version of R was used, and what packages were loaded.
R version 4.4.2 (2024-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Sequoia 15.2
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Los_Angeles
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4
[5] purrr_1.0.2 readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[9] ggplot2_3.5.1 tidyverse_2.0.0 rsvg_2.6.1 DiagrammeRsvg_0.1
[13] DiagrammeR_1.0.11 broom_1.0.7 multilevel_2.7 nlme_3.1-166
[17] mediation_4.5.0 sandwich_3.1-0 mvtnorm_1.2-5 Matrix_1.7-1
[21] MASS_7.3-64 janitor_2.2.1 kableExtra_1.4.0 knitr_1.49
loaded via a namespace (and not attached):
[1] tidyselect_1.2.1 viridisLite_0.4.2 farver_2.1.2 fastmap_1.2.0
[5] digest_0.6.36 rpart_4.1.23 timechange_0.3.0 lifecycle_1.0.4
[9] cluster_2.1.6 magrittr_2.0.3 compiler_4.4.2 rlang_1.1.4
[13] Hmisc_5.2-1 sass_0.4.9 tools_4.4.2 utf8_1.2.4
[17] yaml_2.3.10 data.table_1.15.4 labeling_0.4.3 htmlwidgets_1.6.4
[21] curl_5.2.1 xml2_1.3.6 RColorBrewer_1.1-3 withr_3.0.2
[25] foreign_0.8-87 nnet_7.3-19 grid_4.4.2 fansi_1.0.6
[29] colorspace_2.1-0 scales_1.3.0 cli_3.6.3 crayon_1.5.3
[33] rmarkdown_2.29 generics_0.1.3 rstudioapi_0.16.0 tzdb_0.4.0
[37] visNetwork_2.1.2 minqa_1.2.7 cachem_1.1.0 splines_4.4.2
[41] base64enc_0.1-3 vctrs_0.6.5 V8_5.0.0 boot_1.3-31
[45] jsonlite_1.8.8 bookdown_0.42 hms_1.1.3 Formula_1.2-5
[49] htmlTable_2.4.2 systemfonts_1.1.0 jquerylib_0.1.4 glue_1.8.0
[53] nloptr_2.1.1 stringi_1.8.4 gtable_0.3.5 lme4_1.1-35.5
[57] munsell_0.5.1 pillar_1.9.0 htmltools_0.5.8.1 R6_2.5.1
[61] evaluate_0.24.0 lpSolve_5.6.20 lattice_0.22-6 backports_1.5.0
[65] snakecase_0.11.1 bslib_0.7.0 Rcpp_1.0.13 svglite_2.1.3
[69] gridExtra_2.3 checkmate_2.3.1 xfun_0.49 zoo_1.8-12
[73] pkgconfig_2.0.3