Chapter 17 Linear mixed effects models 1
17.1 Learning goals
- Understanding sources of dependence in data.
- fixed effects vs. random effects.
lmer()syntax in R.- Understanding the
lmer()summary. - Simulating data from an
lmer().
17.2 Load packages and set plotting theme
library("knitr") # for knitting RMarkdown
library("kableExtra") # for making nice tables
library("janitor") # for cleaning column names
library("broom.mixed") # for tidying up linear models
library("patchwork") # for making figure panels
library("lme4") # for linear mixed effects models
library("tidyverse") # for wrangling, plotting, etc. 17.3 Dependence
Let’s generate a data set in which two observations from the same participants are dependent, and then let’s also shuffle this data set to see whether taking into account the dependence in the data matters.
# make example reproducible
set.seed(1)
df.dependence = tibble(participant = 1:20,
condition1 = rnorm(20),
condition2 = condition1 + rnorm(20, mean = 0.2, sd = 0.1)) %>%
mutate(condition2shuffled = sample(condition2)) # shuffles the condition labelLet’s visualize the original and shuffled data set:
df.plot = df.dependence %>%
pivot_longer(cols = -participant,
names_to = "condition",
values_to = "value") %>%
mutate(condition = str_replace(condition, "condition", ""))
p1 = ggplot(data = df.plot %>%
filter(condition != "2shuffled"),
mapping = aes(x = condition, y = value)) +
geom_line(aes(group = participant), alpha = 0.3) +
geom_point() +
stat_summary(fun = "mean",
geom = "point",
shape = 21,
fill = "red",
size = 4) +
labs(title = "original",
tag = "a)")
p2 = ggplot(data = df.plot %>%
filter(condition != "2"),
mapping = aes(x = condition, y = value)) +
geom_line(aes(group = participant), alpha = 0.3) +
geom_point() +
stat_summary(fun = "mean",
geom = "point",
shape = 21,
fill = "red",
size = 4) +
labs(title = "shuffled",
tag = "b)")
p1 + p2 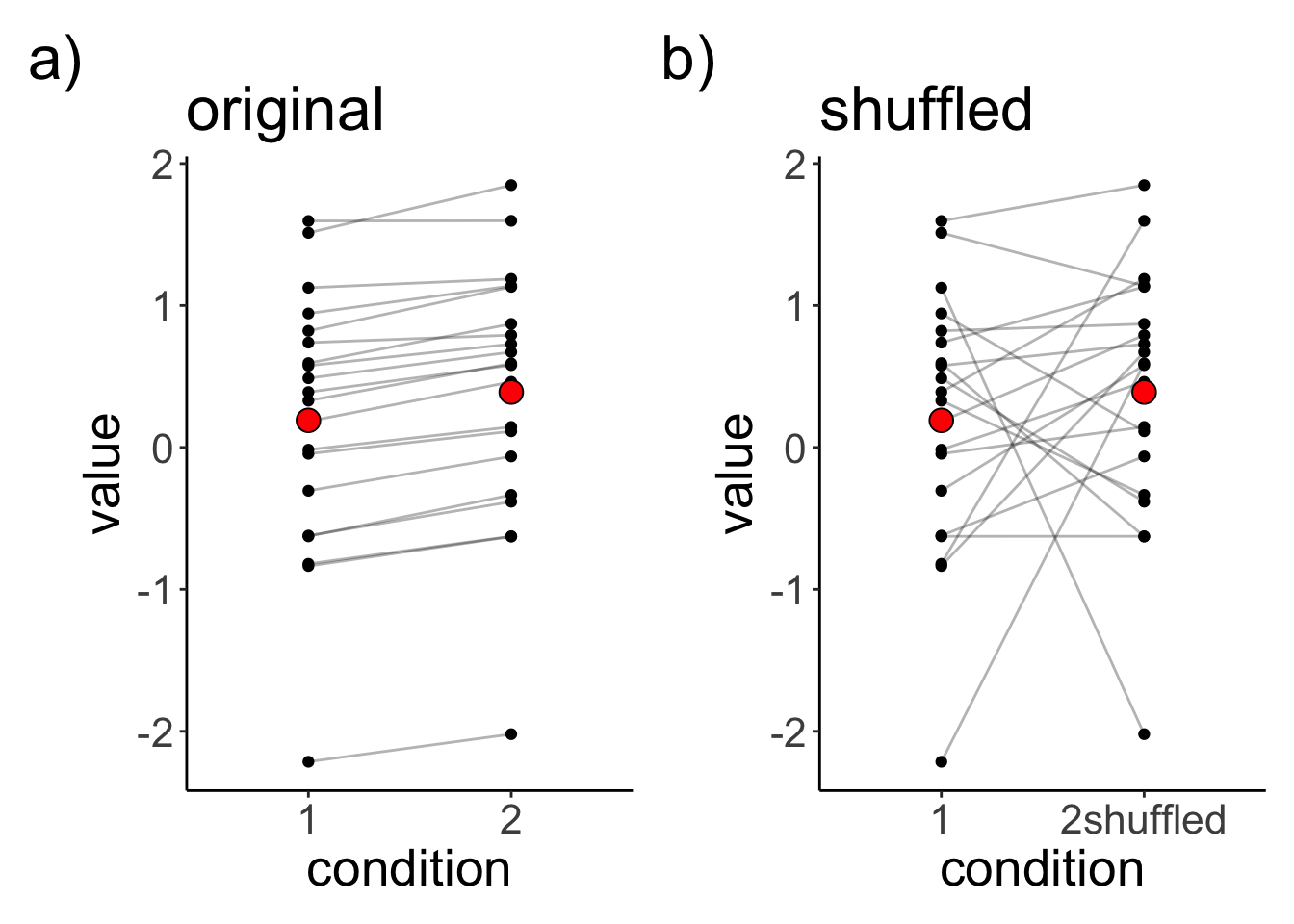
Let’s save the two original and shuffled data set as two separate data sets.
# separate the data sets
df.original = df.dependence %>%
pivot_longer(cols = -participant,
names_to = "condition",
values_to = "value") %>%
mutate(condition = str_replace(condition, "condition", "")) %>%
filter(condition != "2shuffled")
df.shuffled = df.dependence %>%
pivot_longer(cols = -participant,
names_to = "condition",
values_to = "value") %>%
mutate(condition = str_replace(condition, "condition", "")) %>%
filter(condition != "2")Let’s run a linear model, and independent samples t-test on the original data set.
# linear model (assuming independent samples)
lm(formula = value ~ condition,
data = df.original) %>%
summary()
Call:
lm(formula = value ~ condition, data = df.original)
Residuals:
Min 1Q Median 3Q Max
-2.4100 -0.5530 0.1945 0.5685 1.4578
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.1905 0.2025 0.941 0.353
condition2 0.1994 0.2864 0.696 0.491
Residual standard error: 0.9058 on 38 degrees of freedom
Multiple R-squared: 0.01259, Adjusted R-squared: -0.0134
F-statistic: 0.4843 on 1 and 38 DF, p-value: 0.4907t.test(df.original$value[df.original$condition == "1"],
df.original$value[df.original$condition == "2"],
alternative = "two.sided",
paired = F)
Welch Two Sample t-test
data: df.original$value[df.original$condition == "1"] and df.original$value[df.original$condition == "2"]
t = -0.69595, df = 37.99, p-value = 0.4907
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.7792396 0.3805339
sample estimates:
mean of x mean of y
0.1905239 0.3898767 The mean difference between the conditions is extremely small, and non-significant (if we ignore the dependence in the data).
Let’s fit a linear mixed effects model with a random intercept for each participant:
# fit a linear mixed effects model
lmer(formula = value ~ condition + (1 | participant),
data = df.original) %>%
summary()Linear mixed model fit by REML ['lmerMod']
Formula: value ~ condition + (1 | participant)
Data: df.original
REML criterion at convergence: 17.3
Scaled residuals:
Min 1Q Median 3Q Max
-1.55996 -0.36399 -0.03341 0.34400 1.65823
Random effects:
Groups Name Variance Std.Dev.
participant (Intercept) 0.816722 0.90373
Residual 0.003796 0.06161
Number of obs: 40, groups: participant, 20
Fixed effects:
Estimate Std. Error t value
(Intercept) 0.19052 0.20255 0.941
condition2 0.19935 0.01948 10.231
Correlation of Fixed Effects:
(Intr)
condition2 -0.048To test for whether condition is a significant predictor, we need to use our model comparison approach:
# fit models
fit.compact = lmer(formula = value ~ 1 + (1 | participant),
data = df.original)
fit.augmented = lmer(formula = value ~ condition + (1 | participant),
data = df.original)
# compare via Chisq-test
anova(fit.compact, fit.augmented)refitting model(s) with ML (instead of REML)Data: df.original
Models:
fit.compact: value ~ 1 + (1 | participant)
fit.augmented: value ~ condition + (1 | participant)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
fit.compact 3 53.315 58.382 -23.6575 47.315
fit.augmented 4 17.849 24.605 -4.9247 9.849 37.466 1 9.304e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1This result is identical to running a paired samples t-test:
t.test(df.original$value[df.original$condition == "1"],
df.original$value[df.original$condition == "2"],
alternative = "two.sided",
paired = T)
Paired t-test
data: df.original$value[df.original$condition == "1"] and df.original$value[df.original$condition == "2"]
t = -10.231, df = 19, p-value = 3.636e-09
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
-0.2401340 -0.1585717
sample estimates:
mean difference
-0.1993528 But, unlike in the paired samples t-test, the linear mixed effects model explicitly models the variation between participants, and it’s a much more flexible approach for modeling dependence in data.
Let’s fit a linear model and a linear mixed effects model to the original (non-shuffled) data.
# model assuming independence
fit.independent = lm(formula = value ~ 1 + condition,
data = df.original)
# model assuming dependence
fit.dependent = lmer(formula = value ~ 1 + condition + (1 | participant),
data = df.original)Let’s visualize the linear model’s predictions:
# plot with predictions by fit.independent
fit.independent %>%
augment() %>%
bind_cols(df.original %>%
select(participant)) %>%
clean_names() %>%
ggplot(data = .,
mapping = aes(x = condition,
y = value,
group = participant)) +
geom_point(alpha = 0.5) +
geom_line(alpha = 0.5) +
geom_point(aes(y = fitted),
color = "red") +
geom_line(aes(y = fitted),
color = "red")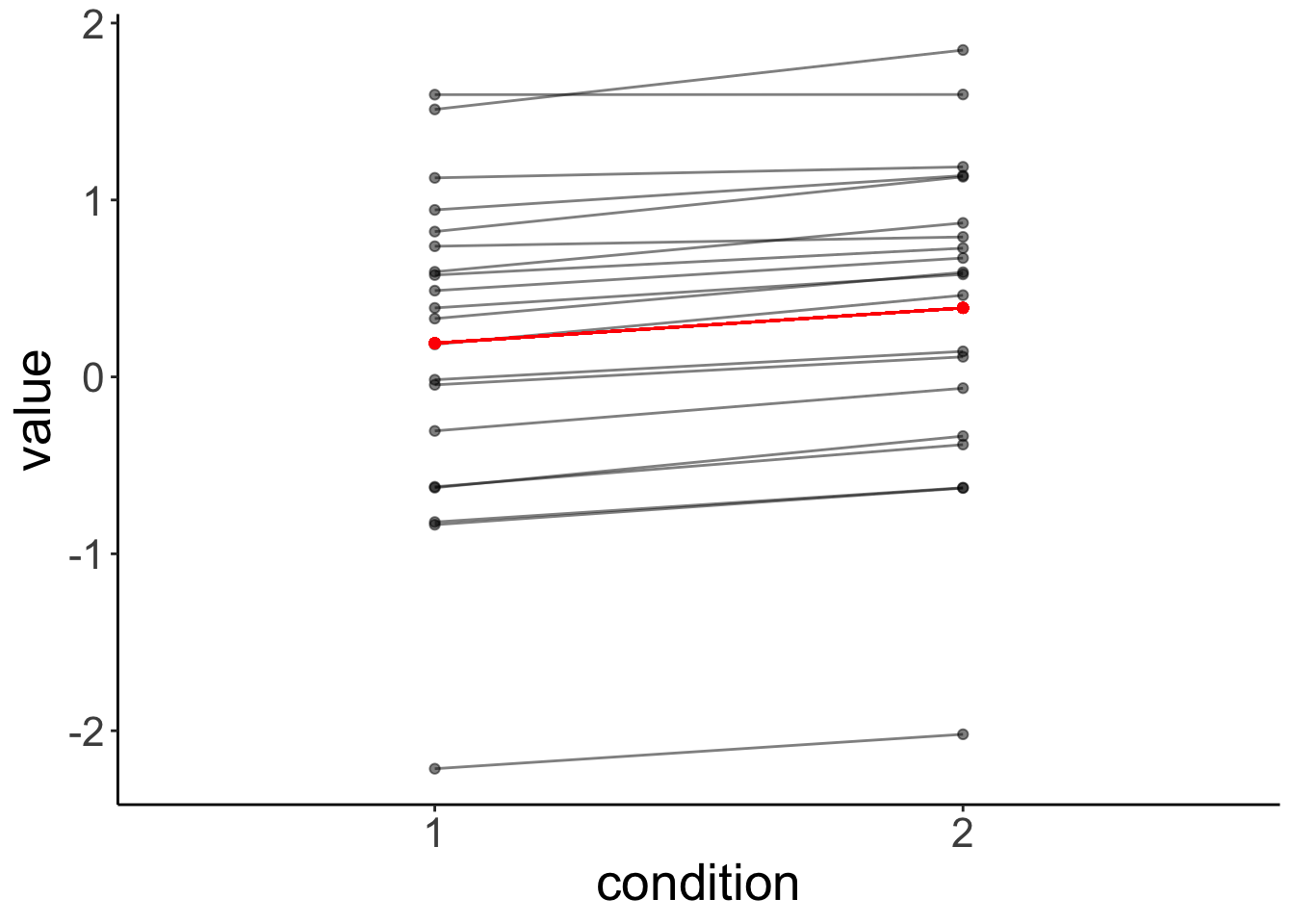
And this is what the residuals look like:
# make example reproducible
set.seed(1)
fit.independent %>%
augment() %>%
bind_cols(df.original %>%
select(participant)) %>%
clean_names() %>%
mutate(index = as.numeric(condition),
index = index + runif(n(), min = -0.3, max = 0.3)) %>%
ggplot(data = .,
mapping = aes(x = index,
y = value,
group = participant,
color = condition)) +
geom_point() +
geom_smooth(method = "lm",
se = F,
formula = "y ~ 1",
aes(group = condition)) +
geom_segment(aes(xend = index,
yend = fitted),
alpha = 0.5) +
scale_color_brewer(palette = "Set1") +
scale_x_continuous(breaks = 1:2,
labels = 1:2) +
labs(x = "condition") +
theme(legend.position = "none")
It’s clear from this residual plot, that fitting two separate lines (or points) is not much better than just fitting one line (or point).
Let’s visualize the predictions of the linear mixed effects model:
# plot with predictions by fit.independent
fit.dependent %>%
augment() %>%
clean_names() %>%
ggplot(data = .,
mapping = aes(x = condition,
y = value,
group = participant)) +
geom_point(alpha = 0.5) +
geom_line(alpha = 0.5) +
geom_point(aes(y = fitted),
color = "red") +
geom_line(aes(y = fitted),
color = "red")
Let’s compare the residuals of the linear model with that of the linear mixed effects model:
# linear model
p1 = fit.independent %>%
augment() %>%
clean_names() %>%
ggplot(data = .,
mapping = aes(x = fitted,
y = resid)) +
geom_point() +
coord_cartesian(ylim = c(-2.5, 2.5))
# linear mixed effects model
p2 = fit.dependent %>%
augment() %>%
clean_names() %>%
ggplot(data = .,
mapping = aes(x = fitted,
y = resid)) +
geom_point() +
coord_cartesian(ylim = c(-2.5, 2.5))
p1 + p2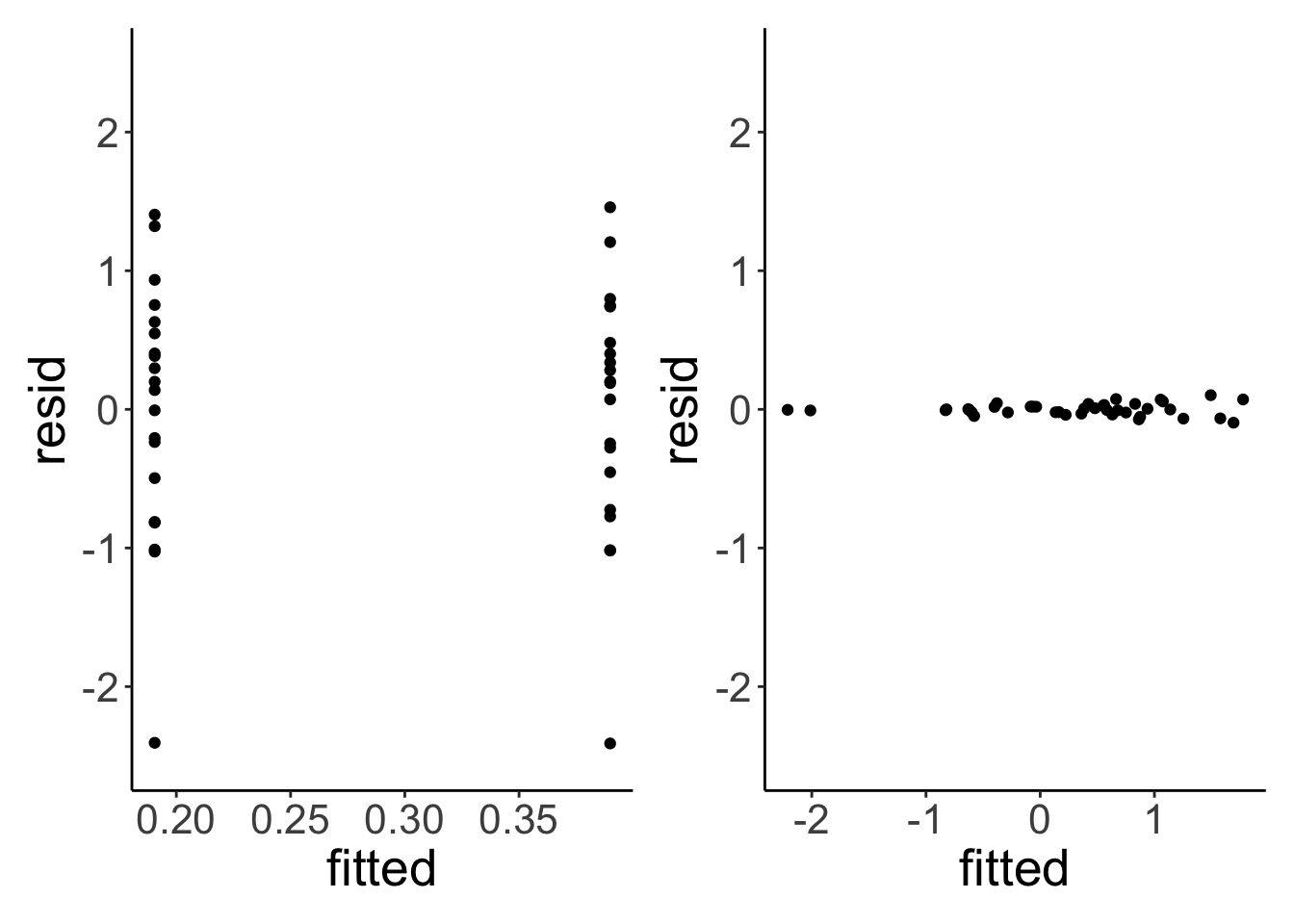
The residuals of the linear mixed effects model are much smaller. Let’s test whether taking the individual variation into account is worth it (statistically speaking).
# fit models (without and with dependence)
fit.compact = lm(formula = value ~ 1 + condition,
data = df.original)
fit.augmented = lmer(formula = value ~ 1 + condition + (1 | participant),
data = df.original)
# compare models
# note: the lmer model has to be entered as the first argument
anova(fit.augmented, fit.compact) refitting model(s) with ML (instead of REML)Data: df.original
Models:
fit.compact: value ~ 1 + condition
fit.augmented: value ~ 1 + condition + (1 | participant)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
fit.compact 3 109.551 114.617 -51.775 103.551
fit.augmented 4 17.849 24.605 -4.925 9.849 93.701 1 < 2.2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Yes, the linear mixed effects model explains the data better than the linear model.
17.5 Session info
Information about this R session including which version of R was used, and what packages were loaded.
R version 4.4.2 (2024-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Sequoia 15.2
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Los_Angeles
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1
[4] dplyr_1.1.4 purrr_1.0.2 readr_2.1.5
[7] tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.1
[10] tidyverse_2.0.0 lme4_1.1-35.5 Matrix_1.7-1
[13] patchwork_1.3.0 broom.mixed_0.2.9.6 janitor_2.2.1
[16] kableExtra_1.4.0 knitr_1.49
loaded via a namespace (and not attached):
[1] gtable_0.3.5 xfun_0.49 bslib_0.7.0 lattice_0.22-6
[5] tzdb_0.4.0 vctrs_0.6.5 tools_4.4.2 generics_0.1.3
[9] parallel_4.4.2 fansi_1.0.6 pkgconfig_2.0.3 RColorBrewer_1.1-3
[13] lifecycle_1.0.4 compiler_4.4.2 farver_2.1.2 munsell_0.5.1
[17] codetools_0.2-20 snakecase_0.11.1 htmltools_0.5.8.1 sass_0.4.9
[21] yaml_2.3.10 nloptr_2.1.1 pillar_1.9.0 furrr_0.3.1
[25] jquerylib_0.1.4 MASS_7.3-64 cachem_1.1.0 boot_1.3-31
[29] nlme_3.1-166 parallelly_1.37.1 tidyselect_1.2.1 digest_0.6.36
[33] stringi_1.8.4 future_1.33.2 bookdown_0.42 listenv_0.9.1
[37] labeling_0.4.3 splines_4.4.2 fastmap_1.2.0 grid_4.4.2
[41] colorspace_2.1-0 cli_3.6.3 magrittr_2.0.3 utf8_1.2.4
[45] broom_1.0.7 withr_3.0.2 scales_1.3.0 backports_1.5.0
[49] timechange_0.3.0 rmarkdown_2.29 globals_0.16.3 hms_1.1.3
[53] evaluate_0.24.0 viridisLite_0.4.2 mgcv_1.9-1 rlang_1.1.4
[57] Rcpp_1.0.13 glue_1.8.0 xml2_1.3.6 minqa_1.2.7
[61] svglite_2.1.3 rstudioapi_0.16.0 jsonlite_1.8.8 R6_2.5.1
[65] systemfonts_1.1.0